Improving the Effectiveness and Efficiency of Stochastic Neighbour Embedding with Isolation Kernel
Abstract
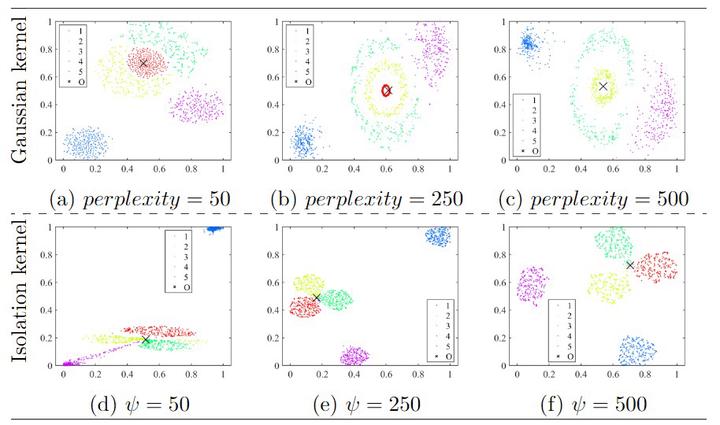
This paper presents a new insight into improving the performance of Stochastic Neighbour Embedding (t-SNE) by using Isolation kernel instead of Gaussian kernel. Isolation kernel outperforms Gaussian kernel in two aspects. First, the use of Isolation kernel in t-SNE overcomes the drawback of misrepresenting some structures in the data, which often occurs when Gaussian kernel is applied in t-SNE. This is because Gaussian kernel determines each local bandwidth based on one local point only, while Isolation kernel is derived directly from the data based on space partitioning. Second, the use of Isolation kernel yields a more efficient similarity computation because data-dependent Isolation kernel has only one parameter that needs to be tuned. In contrast, the use of data-independent Gaussian kernel increases the computational cost by determining n bandwidths for a dataset of n points. As the root cause of these deficiencies in t-SNE is Gaussian kernel, we show that simply replacing Gaussian kernel with Isolation kernel in t-SNE significantly improves the quality of the final visualisation output (without creating misrepresented structures) and removes one key obstacle that prevents t-SNE from processing large datasets. Moreover, Isolation kernel enables t-SNE to deal with large-scale datasets in less runtime without trading off accuracy, unlike existing methods in speeding up t-SNE.